 |
Image Component Library (ICL)
|
 |
Image Component Library (ICL)
|
Generic implementation of D to K dim Self Organizing Map (SOM) More...
#include <SOM.h>

Classes | |
| struct | Neuron |
| SOM internal Neuron struct. More... | |
Public Member Functions | |
| SOM (unsigned int dataDim, const std::vector< unsigned int > &dims, const std::vector< utils::Range< float > > &prototypeBounds, float epsilon=0.1, float sigma=1) | |
| create a new SOM with given data dimension and grid dimensions. More... | |
| ~SOM () | |
| Destructor. More... | |
| void | train (const float *input) |
| trains the net using given input vector and current parameters epsilon and sigma More... | |
| const Neuron & | getWinner (const float *input) const |
| returns a reference of the winner neuron dependent on a given input vector (const) More... | |
| Neuron & | getWinner (const float *input) |
| returns a reference of the winner neuron dependent on a given input vector More... | |
| const std::vector< Neuron > & | getNeurons () const |
| returns the neuron set (const) More... | |
| std::vector< Neuron > & | getNeurons () |
| returns the neuron set More... | |
| const Neuron & | getNeuron (const std::vector< int > &dims) const |
| returns a neuron at given grid location (const) More... | |
| Neuron & | getNeuron (const std::vector< int > &dims) |
| returns a neuron at given grid location More... | |
| unsigned int | getDataDim () const |
| returns the data dimension More... | |
| unsigned int | getSomDim () const |
| returns the SOM dimension (e.g. 2D, 3D ..) More... | |
| const std::vector< unsigned int > | getDimensions () const |
| returns the SOM's grid dimension (e.g. 20x40 for a 2D SOM) More... | |
| void | setEpsilon (float epsilon) |
| sets the current learning rate epsilon More... | |
| void | setSigma (float sigma) |
| sets the current grid distance function standard deviation More... | |
Protected Attributes | |
| unsigned int | m_uiDataDim |
| internal data dimension variable More... | |
| unsigned int | m_uiSomDim |
| internal SOM dimension variable ( = m_vecPrototypeBounds.size() = m_vecDimensions.size() ) More... | |
| std::vector< unsigned int > | m_vecDimensions |
| internal grid dimensions More... | |
| std::vector< utils::Range< float > > | m_vecPrototypeBounds |
| internal bounds for prototype ranges (todo: is it necessary to store them ?) More... | |
| std::vector< Neuron > | m_vecNeurons |
| set of neurons More... | |
| std::vector< unsigned int > | m_vecDimOffsets |
| internal utility offset vector for each dimension More... | |
| float | m_fEpsilon |
| learning rate More... | |
| float | m_fSigma |
| standard deviation for the grid distance function More... | |
Generic implementation of D to K dim Self Organizing Map (SOM)
Input dimension as well as output dimension is freely configurable, i.e. SOM neurons can be aligned in a K dimensional grid with arbitrary cell count for each dimension. For more convenience an inline wrapper class called SOM2D is also available in "iclSOM2D.h"
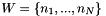
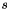
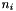 :
: 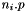 of dim
of dim 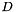
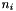 :
: 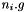 of dim
of dim 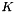
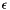
new input vector: 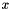
![\[ \forall r \in \{1,...,n\} \\ \Delta n_r.p = \epsilon \cdot h(n_r.g,h_s.g) \cdot (x-n_r.p) \]](form_70.png)
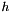 )
)
![\[ h(a,b) = e^{-\frac{(a-b)^2}{s\sigma^2}} \]](form_72.png)
| icl::math::SOM::SOM | ( | unsigned int | dataDim, |
| const std::vector< unsigned int > & | dims, | ||
| const std::vector< utils::Range< float > > & | prototypeBounds, | ||
| float | epsilon = 0.1, |
||
| float | sigma = 1 |
||
| ) |
create a new SOM with given data dimension and grid dimensions.
| dataDim | dimension of data elements and prototype vectors |
| dims | grid dimension array. dims.size() defines the SOM'S grid dimension. dims[i] defines the cell count for dimension i. E.g. to create a 2D 40x40 SOM grid dims has to be 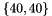 . . |
| prototypeBounds | Internals for new randomly created prototype vector elements. prototypeBounds.size() must be equal to dataDim. E.g. to initialize a SOM with prototypes randomly distributed in a dataDim-D unity cube ![$[-1,1]^{dataDim}$](form_74.png) , prototypeBounds must be , prototypeBounds must be 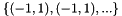 (where the ranges are written as tuples). (where the ranges are written as tuples). |
| epsilon | initial learning rate |
| sigma | initial standard deviation for the grid distance function 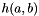 |
| icl::math::SOM::~SOM | ( | ) |
Destructor.
|
inline |
returns the data dimension
|
inline |
| const Neuron& icl::math::SOM::getNeuron | ( | const std::vector< int > & | dims | ) | const |
returns a neuron at given grid location (const)
| Neuron& icl::math::SOM::getNeuron | ( | const std::vector< int > & | dims | ) |
returns a neuron at given grid location
| const std::vector<Neuron>& icl::math::SOM::getNeurons | ( | ) | const |
returns the neuron set (const)
| std::vector<Neuron>& icl::math::SOM::getNeurons | ( | ) |
returns the neuron set
|
inline |
returns the SOM dimension (e.g. 2D, 3D ..)
| const Neuron& icl::math::SOM::getWinner | ( | const float * | input | ) | const |
returns a reference of the winner neuron dependent on a given input vector (const)
| Neuron& icl::math::SOM::getWinner | ( | const float * | input | ) |
returns a reference of the winner neuron dependent on a given input vector
| void icl::math::SOM::setEpsilon | ( | float | epsilon | ) |
sets the current learning rate epsilon
| void icl::math::SOM::setSigma | ( | float | sigma | ) |
sets the current grid distance function standard deviation
| void icl::math::SOM::train | ( | const float * | input | ) |
trains the net using given input vector and current parameters epsilon and sigma
|
protected |
learning rate
|
protected |
standard deviation for the grid distance function
|
protected |
internal data dimension variable
|
protected |
internal SOM dimension variable ( = m_vecPrototypeBounds.size() = m_vecDimensions.size() )
|
protected |
internal grid dimensions
|
protected |
internal utility offset vector for each dimension
|
protected |
set of neurons
|
protected |
internal bounds for prototype ranges (todo: is it necessary to store them ?)
 1.8.15
1.8.15