|
| | SOM2D (unsigned int dataDim, unsigned int nXCells, unsigned int nYCells, const std::vector< utils::Range< float > > &prototypeBounds, float epsilon=0.1, float sigma=1) |
| | Wrapper constructor for 2D SOMs. More...
|
| |
| const Neuron & | getNeuron (int x, int y) const |
| | Wrapper function to access a neuron at a certain grid position. More...
|
| |
| | SOM (unsigned int dataDim, const std::vector< unsigned int > &dims, const std::vector< utils::Range< float > > &prototypeBounds, float epsilon=0.1, float sigma=1) |
| | create a new SOM with given data dimension and grid dimensions. More...
|
| |
| | ~SOM () |
| | Destructor. More...
|
| |
| void | train (const float *input) |
| | trains the net using given input vector and current parameters epsilon and sigma More...
|
| |
| const Neuron & | getWinner (const float *input) const |
| | returns a reference of the winner neuron dependent on a given input vector (const) More...
|
| |
| Neuron & | getWinner (const float *input) |
| | returns a reference of the winner neuron dependent on a given input vector More...
|
| |
| const std::vector< Neuron > & | getNeurons () const |
| | returns the neuron set (const) More...
|
| |
| std::vector< Neuron > & | getNeurons () |
| | returns the neuron set More...
|
| |
| const Neuron & | getNeuron (const std::vector< int > &dims) const |
| | returns a neuron at given grid location (const) More...
|
| |
| Neuron & | getNeuron (const std::vector< int > &dims) |
| | returns a neuron at given grid location More...
|
| |
| unsigned int | getDataDim () const |
| | returns the data dimension More...
|
| |
| unsigned int | getSomDim () const |
| | returns the SOM dimension (e.g. 2D, 3D ..) More...
|
| |
| const std::vector< unsigned int > | getDimensions () const |
| | returns the SOM's grid dimension (e.g. 20x40 for a 2D SOM) More...
|
| |
| void | setEpsilon (float epsilon) |
| | sets the current learning rate epsilon More...
|
| |
| void | setSigma (float sigma) |
| | sets the current grid distance function standard deviation More...
|
| |
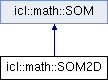
Simple Wrapper class of the generic SOM Implementation for 2D SOMs.
As the SOM class provides a very abstract interface for generic K-D Self Organizing Maps, the SOM2D class offers a more convenient interface for exactly 2D SOMs.
Therefore it has a specialized constructor (now receiving the count of x-Cells and y-Cells directly as well as a specialized getNeuron()- function.
 Public Member Functions inherited from icl::math::SOM
Public Member Functions inherited from icl::math::SOM Protected Attributes inherited from icl::math::SOM
Protected Attributes inherited from icl::math::SOM 1.8.15
1.8.15